
Characterization of the malignant cells and microenvironment of infantile fibrosarcoma via single-cell RNA sequencing
Highlight box
Key findings
• We clarified the single-cell transcriptional characteristics of infantile fibrosarcoma (IFS) and identified novel targets for diagnosis and therapy.
What is known and what is new?
• Although it has been partly associated with ETV6-NTRK3 gene fusion, IFS has no specific immunohistochemical diagnostic markers. Moreover, no universal characteristics of the tumor environment within IFS have been identified.
• POSTN, IGFBP2 and CTHRC1 were found to be potential novel markers for IFS, endothelial cells and M2-like macrophages play a primary role in the immune microenvironment of IFS.
What is the implication, and what should change now?
• This study represents an opportunity for making clinical advancements in the immunohistochemical diagnosis and treatment of IFS. Further exploration should be conducted to validate these findings.
Introduction
Infantile fibrosarcoma (IFS) is a rare type of nonrhabdomyosarcoma soft tissue tumor and consists of malignant spindled fibroblasts originating from mesenchymal cells. It is the most common soft tissue sarcoma in children under 1 year old, with a higher incidence in males than in females, and is often located in the distal extremities (1). The tumor typically exhibits widespread positivity for vimentin but is negative for desmin and S100 protein (2,3). Nonetheless, specific tumor markers have not been established for immunohistochemical diagnosis. Moreover, IFS demonstrates rapid growth, resulting in tumor diameters larger than 5 cm at the onset, with potential involvement of critical blood vessels and nerves (1). This significantly complicates the possibility of radical surgical resection, which is the primary therapeutic intervention for IFS, and places the patient at a considerable risk of disability or mutilation.
In 1998, Knezevich et al. first reported the chromosomal translocation t(12;15) (p13;q25) in children with IFS, which results in ETV6-NTRK3 gene fusion, and linked it to oncogenesis (4). Subsequently, other NTRK gene-related chromosomal translocations were also identified in IFS (5-7). These studies have explored the genetic characteristics of IFS at the DNA level to improve the diagnosis and treatment of the disease. Researchers have attempted to apply NTRK-targeted drugs, such as larotrectinib, for treating IFS and achieved promising results in clinical trials (8-10). However, NTRK gene-related chromosomal translocations have been confirmed to be relevant to other tumors, and other NTRK gene-unrelated chromosomal variations, such as BRAF rearrangements, have been reported in IFS (1,5). This suggests that this diagnostic approach, despite its high cost, may lack specificity and universality to some extent. Furthermore, the related targeted therapies also face certain challenges, including tumor resistance, adverse reactions, and high costs (8).
The tumor microenvironment (TME), which directly influences cancer cells and plays a crucial role in cancer development and progression, has become a focal point of breakthrough research in emerging therapeutic strategies. Regarding IFS, Zhu et al. have utilized immunohistochemistry and multicolor flow cytometry to prove that IFS tumors are highly immunogenic (11). They propose that the expansion of tumor-infiltrating lymphocytes followed by adoptive cell transfer could be a potential immunotherapy for IFS patients. Yet, the overall and precise characteristics of the TME, as well as the interactions between malignant cells and other cells within IFS, remain largely unknown.
In recent years, single-cell RNA sequencing (scRNA-seq) has emerged as a robust analytical technique that allows for the investigation of omics information at the individual cell level (12). It provides unprecedented resolution for comprehensively understanding the cellular diversity within complex cancers. Through comparative analysis with normal cells, scRNA-seq can discern malignant cells, characterize distinct malignant cell populations, and elucidate their shared or unique gene expression profiles and functions (13). This advancement holds promise for prognostic prediction and targeted therapy development. Furthermore, scRNA-seq facilitates the investigation of the TME and the interactions among its cellular constituents, providing valuable insights for targeted immunotherapy (14).
We therefore conducted scRNA-seq on lesion tissues from patients with IFS to reveal the universal transcriptional characteristics of both the malignant cells and the TME within this disease, and identify novel targets for immunohistochemical diagnosis, which demands lower cost, and treatment. We present this article in accordance with the STREGA reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-24-66/rc).
Methods
Patient and sample collection
This study performed scRNA-seq for fresh samples from four patients, who were diagnosed with IFS and received biopsy or radical surgery at the Department of Oncology from the Children’s Hospital of Fudan University. Besides, samples from additional 19 patients were collected for histopathological examinations. Prior to surgery, none of the patients had undergone chemotherapy, radiotherapy, or targeted therapy. This study was approved by the Medical Ethics Committee of the Children’s Hospital of Fudan University [approval No. (2020). 419] and conducted in accordance with the Declaration of Helsinki (as revised in 2013). Prior to participation, informed written consent was obtained from the parents of the 23 patients.
Healthy donor data
Healthy control samples were obtained from a deceased brain-dead donor during clinical transplantation. The donor was confirmed to be free from chronic diseases, infections, and malignancies. Tissues from 15 different organs were collected and separately sequenced, including the blood, bone marrow, liver, bile duct, lymph nodes (hepatic hilum and mesentery), spleen, heart (apex), bladder, trachea, esophagus, stomach, small intestine, rectum, skin, and muscle (thigh). The dataset was sourced from the Gene Expression Omnibus (GEO) with accession number GSE159929 (15).
Generation of scRNA-seq data
Samples were processed within an hour after surgery, adhering to the established experimental techniques using the Chromium Single Cell 3’ Library V2/V3 kit (10x Genomics, Pleasanton, CA, USA). Lesion tissues were enzymatically digested and underwent density gradient centrifugation with lymphocyte separation medium for mononuclear cells, which were loaded into a Chromium Controller (10x Genomics). The resulting barcoded complementary DNAs (cDNAs) were used for library construction. Raw data were converted into FASTQs format via Illumina bcl2fastq software (Illumina, San Diego, CA, USA), followed by alignment to the human genome (GRCh38) utilizing the CellRanger v. 3.0.1 (10x Genomics) pipeline in accordance with the manufacturer’s instructions. Subsequently, a digital gene-cell matrix was generated from this processed data.
Single-cell data quality control and processing
Seurat v4 was employed to process and analyze the gene expression matrix (16). The scRNA-seq data from healthy donors underwent identical processing steps. Cells were filtered based on gene expression, removing those with fewer than 500 or more than 6,000 detected genes to ensure data quality. Cells exhibiting over 10% gene expression derived from the mitochondrial genome were excluded. Nuclear mitochondrial and ribosomal genes were omitted from subsequent analyses. Additionally, Scrublet was used to remove cell doublets (17).
After quality control, the data underwent normalization utilizing the SCTransform function. Subsequently, highly variable genes were identified and examined with FindVariableFeatures function. Principal component analysis (PCA) was conducted using the RunPCA function, and clusters were identified through the FindNeighbors and FindClusters functions. The visualization was realized respectively via t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) using the RunTSNE and RunUMAP functions. Differentially expressed genes (DEGs) of each cluster were determined with the FindAllMarkers function.
Single-cell data integration
The data separately generated from 14,879, 15,572, 10,142, and 9,143 isolated cells from four lesion tissues of IFS and 18,405 cells from healthy donor were merged for integration analysis. The Harmony algorithm was employed to rectify technical batch effects within the merged dataset (18).
Cluster definition
Based on the DEGs and conventional cell markers, cell types were manually assigned to the clusters. Markers used to type cells included KRT14, KRT1, DMKN (epithelial cells), PECAM1, VWF, PLAVP (endothelial cells), ACTA2, MYL9, TAGLN (smooth muscle cells), MMP9, COL1A1 (fibroblasts), MMP2, ACTA2, COL1A1 (myofibroblasts), PTPRC (immune cells), CD2, CD3D, CD3E (T cells), GZMK, GZMA, GZMB, GZMH (effector T cells), IL2RA, FOXP3, IKZF2 [regulatory T cells (Tregs)], TRGV9, TRDV2 (γδ T cells), NKG7, NCAM1, KLRD1 [natural killer (NK)/natural killer T (NKT) cell], C1QA, C1QB, MSR1 (macrophages), S100A8, S100A9, VCAN (monocytes), CD1C (classical dendritic cells), IRF8, CLEC4C (plasmacytoid dendritic cells), CD19, CD79A, SDC1, XBP1 (plasma cells), KIT, TPSAB1, and TPSB2 (mast cells) (15,19,20).
Inferring copy number variation from scRNA-seq (InferCNV) analysis
InferCNV, based on the hidden Markov model (InferCNV of the Trinity CTAT Project; https://github.com/broadinstitute/inferCNV), was employed to infer the copy number variations (CNVs) in malignant-like cells. Normal cells from healthy donors were used as reference. The analysis procedure followed guidelines outlined in the tutorial (https://github.com/broadinstitute/inferCNV/wiki).
Specific marker identification for malignant cells
The Wilcoxon rank-sum test was employed to assess and characterize DEGs identified by the FindAllMarkers function. The novel markers were identified using a default threshold of two for average fold change and a filter for the minimum delta percent of cells [(percentage of cluster1 – percentage of cluster2)/percentage of cluster1 ×100%] greater than 90% (19).
Pearson correlation calculation
Scanpy (21), a scalable toolkit for analyzing single-cell gene expression data (https://github.com/scverse/scanpy), was used to calculate the Pearson correlation of transcriptional features among malignant cells for hierarchical clustering and potential cell subtype visualization using the sc.pl.correlation_matrix function.
Cell function
Gene functional scoring of macrophages was performed using the AddModuleScore algorithm according to the M1-like and M2-like reference sets (22). The top 50 DEGs of each cell type were selected for Gene Ontology (GO) analysis. This analysis aimed to explore the functional status of the cells and was conducted utilizing the clusterProfiler package (an R package for comparing biological themes among gene clusters; https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html).
Cell-cell and ligand-receptor interactions
CellPhoneDB, a public repository of ligands, receptors and their interactions, was used for cell-cell and ligand-receptor interaction inference in the IFS microenvironment (23). We established a lower threshold for the expression proportion of any ligand or receptor within each cell type at 10%. Additionally, the number of permutations was set to 1,000.
Hematoxylin and eosin (HE) staining and immunohistochemistry
The formalin-fixed and paraffin-embedded tissue was cut into 4-mm-thick sections and affixed onto the slides, which were subjected to HE staining and immunohistochemistry. The DAB polymer detection kit (GeneTech, Shanghai, China) was used according to the manufacturer’s provided protocol, and Tris-EDTA (pH 9.0) buffer was used for antigen retrieval. Anti-CTHRC1 antibody (1:800; Ab256458, Abcam, Cambridge, UK), anti-periostin antibody (1:100; Ab215199, Abcam), and anti-IGFBP2 antibody (1:250, Ab188200, Abcam) were employed.
Results
Clinical characteristics of the patients with IFS
Table 1 presents a summary of the clinical characteristics observed in the four patients diagnosed with IFS. Three of the patients exhibited various clinical manifestations within 24 hours after birth, including vomiting, right eye protrusion, and bilateral lower limb swelling. The fourth patient was found to have a painless massive lump in the right hip at the age of 2 months. All patients were subjected to biopsy or surgical resection. Prior to the surgery, the patients underwent comprehensive evaluations, primarily with computed tomography (CT) and isotopic bone imaging, and no regional lymph node infiltration or distant metastasis was identified. Pathological diagnosis of the four patients’ samples revealed the presence of IFS and the identification of ETV6-NTRK3 gene fusion.
Table 1
| Patient ID | Gender | Age at onset | Clinical manifestations | Local lesion | Other lesions | Surgery | Histopathological characteristics | ETV6-NTRK3 gene fusion | IRS staging | Follow-up | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | Diameter at diagnosis (cm) | Diagnosis | IHC | |||||||||
| Patient 1 | M | After birth | Vomit | Abdominal and pelvic cavity | 8.80 | No | Biopsy | IFS | Vimentin (+), Ki-67 30% (+), alpha-SMA (−), CD34 (−), S-100 (−), Desmin (−) |
Yes | III | Dead |
| Patient 2 | M | 2 months old | Painless lump | From the pelvic area to the right buttock | 1.30 | No | Biopsy | IFS | Vimentin (+), Ki-67 40% (+), alpha-SMA (+), CD34 (−), S-100 (−), Desmin (−) |
Yes | III | Survival |
| Patient 3 | F | After birth | Right eye protrusion | Behind the right eyeball | 3.50 | No | Resection | IFS | Vimentin (+), Ki-67 20% (+), alpha-SMA (−), CD34 (−), S-100 (−), Desmin (−) |
Yes | II | Survival |
| Patient 4 | M | After birth | Vomit and bilateral lower limb swelling | From the pelvic area to the right buttock | 6.00 | No | Biopsy | IFS | Vimentin (+), Ki-67 15% (+), alpha-SMA (+), CD34 (−), S-100 (+), Desmin (−) |
Yes | III | Survival |
IFS, infantile fibrosarcoma; M, male; F, female; cm, centimeter; IHC, immunohistochemistry; SMA, smooth muscle action; IRS, intergroup rhabdomyosarcoma study.
Patient 1 succumbed to the disease within 1 month after the biopsy. Patient 2 discontinued treatment after completing four cycles of chemotherapy due to financial constraints and is currently living with both old and new lesions. Patient 3 experienced local recurrence 2 months after resection, achieved complete remission after taking larotrectinib for 4 months, and continued taking the drug for an additional 12 months; upon cessation of larotrectinib, no recurrence or metastasis occurred. Patient 4 also took larotrectinib after biopsy surgery and obtained partial remission according to the magnetic resonance imaging (MRI) examination performed 7 months later.
Single-cell atlas of IFS
We performed scRNA-seq on tumor specimens obtained from four patients with IFS and obtained four transcriptomic datasets, comprising 14,879, 15,572, 10,142, and 9,143 cells, respectively. Additionally, we acquired the transcriptomic data from a public database (GSE159929), comprising 13,070 fibroblasts, 2,090 myofibroblasts, and 3,245 smooth muscle cells for cell identification (15). In total, we identified 14 distinct cell populations (Figure 1A) according to the known markers (see Methods for details), including epithelial cells, endothelial cells, normal smooth muscle cells, normal fibroblasts, normal myofibroblasts, classical dendritic cells, plasmacytoid dendritic cells, plasma cells, mast cells, macrophages, monocytes, T cells, NK/NKT cells, and malignant-like cells (Figure 1B-1E).
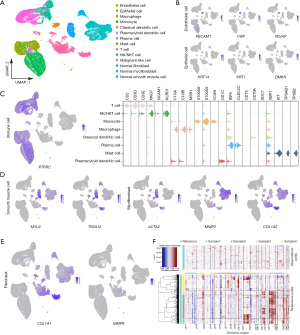
A total of 11,663 malignant-like cells were clustered independently and were identified with specific fibroblast markers (MMP9, COL1A1). Some of them also expressed the myofibroblast marker ACTA2. Large-scale CNVs were inferred from the transcriptomes to distinguish malignant from nonmalignant cells (Figure 1F). We found that nearly all the identified malignant-like cells exhibited gains in 1p, 3p, and 20p when compared to normal cells, affirming their malignant nature.
Single-cell characteristics of malignant cells in IFS
To identify potential novel markers or therapeutic targets for IFS, we conducted differential gene expression analysis by comparing the total malignant cells of four samples with normal cells from the public database. In all, we identified 1,249 DEGs. Among them, we identified three protein-coding genes (POSTN, IGFBP2, and CTHRC1) as potential novel markers for IFS, which were expressed in over 50% of malignant cells and exhibited an average fold change >1.5 and delta percentage >90% compared to normal cells (Figure 2A) (19).
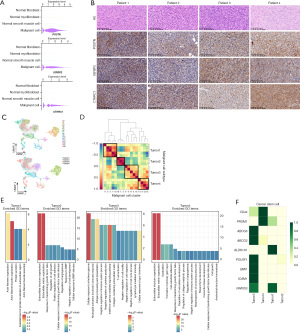
Next, we conducted immunohistochemical staining to examine their expression in IFS tissues (samples from 23 patients, including four scRNA-seq samples and additional 19 samples unsequenced). As expected, these genes were not only universally expressed in the four sequenced lesion tissues (Figure 2B), but also in the validation cohort (POSTN expressed in 18/19 samples, IGFBP2 in 17/19 samples, and CTHRC1 in 18/19 samples). Additionally, we examined previously reported IFS-specific ETV6-NTRK3 gene fusion in IFS, which were identified in 11 of 23 patients (Table 2). This suggests that POSTN, IGFBP2, and CTHRC1 may serve as novel markers for IFS.
Table 2
| Patient ID | Gender | Age at onset | Clinical manifestations | Local lesion | Surgery | Diagnosis | IHC | ETV6-NTRK3 gene fusion | IRS staging | Follow-up | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | Diameter at diagnosis (cm) | CTHRC1 | POSTN | IGFBP2 | |||||||||
| Patient 4 | M | After birth | Painless lump | Head | 9.38 | Resection | IFS | (+) | (+) | (+) | Yes | I | Survival |
| Patient 5 | M | After birth | Painless lump | Head | 19.00 | Resection | IFS | (+) | (+) | (+) | No | I | Survival |
| Patient 6 | F | 2 months old | Painless lump | From the abdomen to the back | 8.92 | Resection | IFS | (+) | (+) | (+) | No | III | Survival |
| Patient 7 | M | After birth | Painless lump | From left shoulder joint to the back | 5.40 | Resection | IFS | (+) | (+) | (−) | No | III | Survival |
| Patient 8 | M | After birth | Painless lump | Head | 1.73 | Resection | IFS | (+) | (+) | (+) | No | I | Survival |
| Patient 9 | F | 7 days old | Painless lump | Right thigh | 5.60 | Biopsy | IFS | (+) | (+) | (+) | No | III | Survival |
| Patient 10 | F | 19 months old | Painless lump | Back | 2.14 | Resection | IFS | (+) | (+) | (+) | No | I | Survival |
| Patient 11 | F | 5 months old | Melena | Abdominal cavity | 3.50 | Resection | IFS | (+) | (+) | (−) | No | II | Survival |
| Patient 12 | F | 12 months old | Painless lump | Left foot | 5.29 | Resection | IFS | (-) | (+) | (+) | No | III | Survival |
| Patient 13 | M | 7 days old | Abdominal distention | Abdominal cavity | 8.55 | Resection | IFS | (+) | (+) | (+) | No | I | Survival |
| Patient 14 | F | After birth | Painless lump | Left calf | 5.60 | Biopsy | IFS | (+) | (+) | (+) | Yes | III | Survival |
| Patient 15 | M | After birth | Painless lump | Abdominal cavity | 6.60 | Resection | IFS | (+) | (+) | (+) | Yes | I | Survival |
| Patient 16 | F | 3 days old | Painless lump | Right thigh | 5.50 | Biopsy | IFS | (+) | (+) | (+) | Yes | IV (lung) | Survival |
| Patient 17 | M | 3 days old | Painless lump | Right thigh | 7.31 | Resection | IFS | (+) | (+) | (+) | No | I | Survival |
| Patient 18 | M | After birth | Painless lump | Right chest wall | 12.60 | Resection | IFS | (+) | (+) | (+) | No | II | Survival |
| Patient 19 | M | After birth | Painless lump | From the abdomen to the back | 5.60 | Resection | IFS | (+) | (−) | (+) | Yes | III | Survival |
| Patient 20 | M | 4 days old | Fever, vomit and poor appetite | Abdominal cavity | 2.50 | Resection | IFS | (+) | (+) | (+) | Yes | I | Survival |
| Patient 21 | M | 3 months old | Painless lump | Right thigh | 4.81 | Resection | IFS | (+) | (+) | (+) | Yes | III | Survival |
| Patient 22 | M | After birth | Abdominal distension and genitalia swelling | Abdominal cavity | 13.80 | Biopsy | IFS | (+) | (+) | (+) | No | III | Dead |
IFS, infantile fibrosarcoma; M, male; F, female; cm, centimeter; IHC, immunohistochemistry; IRS, intergroup rhabdomyosarcoma study.
Moreover, we isolated and analyzed malignant cells separately to explore intertumoral heterogeneity. Sixteen malignant cell clusters were obtained (Figure 2C). According to the Pearson correlation as calculated with Scanpy (21), these clusters could be classified into four subtypes (Figure 2C,2D). We selected the top 50 DEGs for each subtype for GO analysis (Figure 2E). Some DEGs in the three subtypes (tumor 2, tumor 3, and tumor 4) were enriched in common pathways of extracellular matrix organization. Additionally, the DEGs of tumor 2 were enriched in cell growth and adhesion, those of tumor 3 were enriched in immune response, and those of tumor 4 were related to embryonic mesenchymal development. DEGs of tumor 1 were associated with actin filament organization, indicating a potential phenotype of myofibroblast-like malignant cells. This suggests different functional characteristics of various malignant cell subtypes.
Besides, we evaluated the cancer stemness of various malignant cell subtypes based on cancer stem cell (CSC) biomarkers (24,25), including CD44, PROM1, ABC family, ALDH1, POU5F1, BMI1, ICAM1 and NANOG (Figure 2F). Our findings indicated the presence of CSC phenotypes in tumor 1, tumor 3, and tumor 4.
The TME of IFS at single-cell resolution
To further characterize the TME of IFS, we further annotated T cells and macrophages based on conventional cell markers (Figure 3A). Five subtypes of T cells were identified, including CD4+ effector T cells, CD8+GZMK+ effector T cells, CD8+GZMK− effector T cells, Tregs, and γδ T cells (Figure 3B). Meanwhile, AddModuleScore was employed to assess the function of macrophages in IFS based on the known gene sets (22). The majority of cells attained higher scores of the alternatively activated M2-like macrophages compared to the classically activated M1-like macrophages (t-test; P<0.05), indicating a tendency for macrophages to primarily exert anti-inflammatory functions and promote tumor development in IFS (Figure 3C).
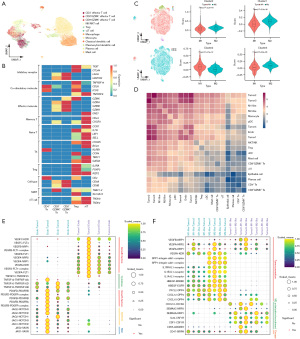
We predicted the cell-cell communication networks in IFS based on CellPhoneDB (23) to explore the intrinsic connections of IFS (Figure 3D). The interactions observed between malignant cells were notably more prevalent with endothelial cells and macrophages, compared with epithelial cells, dendritic cells, plasma cells, mast cells, monocytes, T cells, and NK/NKT cells. Notably, endothelial cells and macrophages emerged as prominent participants in the intricate cell-cell communication network within this microenvironment, indicating their potential pivotal roles in cell-cell interactions among cells in IFS.
We identified multiple interesting receptor-ligand interactions that might contribute to tumor progression. Angiocrine factors were involved between malignant cells and endothelial cells (26), including growth factors (VEDGF and PDGF), cytokines (TNF superfamily), and other factors like NOTCH and Slit2 (Figure 3E). Meanwhile, we found M2-like macrophages might promote tumor progression via VEGFA-NRP1/NRP2, SPP1-integrin, IL1B-IL1R, HBEGF-ERBB2/EGFR, and CXCL2/CXCL10/CXCL12-DPP4 interactions (Figure 3F) (27-34). Additionally, malignant cells might reduce M1-like macrophage infiltration and enhance M2-like macrophage recruitment and polarization by tumor-immune interactions such as VEGFA/SEMA3A/SEMA3C-NRP1/NRP2, CXCL12-CXCR4, IL-34/CSF1-CSF1R, and CD55-ADGRE5 (Figure 3F) (28,31,35-39). The CD47-SIRPA pathway signaling ‘do not eat me’ might enhance immune escape of CD47+ malignant cells (Figure 3F) (31).
Discussion
Although less malignant compared to adult fibrosarcoma, IFS has a high degree of local invasiveness (40). Tumors often present with large volumes and surround vital blood vessels and nerves, thereby increasing the risk of functional impairment associated with radical surgical resection. The early diagnosis and treatment of IFS are critical to patient treatment. However, the pathological diagnosis of IFS relies on characteristic cellular arrangements and lacks specific immunohistochemical markers (3). When patients with IFS lacking these distinctive cellular arrangements, it becomes challenging to differentiate IFS from other spindle cell tumors. Recently, the discovery of the ETV6-NTRK3 fusion gene has provided some assistance in IFS diagnosis and treatment, but its sensitivity and specificity still require further consideration (8). As demonstrated in our study, only 11 of 23 (47.83%) patients with IFS were tested positive for the ETV6-NTRK3 fusion gene. Moreover, this gene fusion has also been identified in other tumors, such as congenital mesoblastic nephroma (5). Novel specific markers for the immunohistochemical diagnosis and treatment of IFS are crucially needed.
Recently, TME, which directly influences cancer cells and plays a crucial role in cancer development and progression, has emerged as a rapidly expanding area of interest. A better understanding of the structural and the functional characteristics of TME provides insights into potential novel therapeutic targets, including immunotherapies and anti-angiogenic therapies (41). However, the comprehensive characteristics of the TME within IFS is unclear.
In this study, we conducted scRNA-seq on lesion tissues from patients with IFS and explored the molecular traits of IFS compared to normal cells from healthy donors. This is the first study to characterize and summarize the malignant cells and TME of IFS at a single-cell resolution. We distinguished malignant cells of IFS from normal fibroblasts, myofibroblasts, and smooth muscle cells, and identified three genes (POSTN, IGFBP2, and CTHRC1) that were explicitly expressed in them and thus may be potential novel markers for IFS.
POSTN encodes a secreted cell adhesion protein that promotes cell survival, invasion, angiogenesis, and epithelial-mesenchymal transition (EMT) in the oncogenesis of various adult malignancies, such as colon cancer and ovarian cancer (42,43). POSTN is highly expressed in IFS malignant cells (average fold change =2.73, delta percentage =95%), suggesting its potential role in IFS. IGFBP2, an encoding gene for a secreted protein or cytoplasmic signaling effector protein, has been identified as an important oncogene associated with malignant cell proliferation, invasion, and EMT (44). It is currently considered a valuable biomarker for patient diagnosis and prognosis as well as a potential therapeutic target for malignant diseases (45). Additionally, CTHRC1 encodes an extracellular matrix protein that is involved in tissue remodeling and immune reactions (46). Its high expression has been confirmed in various adult cancers, such as gastric cancer and hepatocellular carcinoma, and it plays a role in tumorigenesis by upregulating the signaling pathways of TGF-β, PI3K, and Wnt (47). Given these factors, it is worth exploring the role of IGFBP2 and CTHRC1 in IFS.
Moreover, we identified four malignant cell subtypes with distinct functions denoted as tumor 1, tumor 2, tumor 3, and tumor 4. According to the GO analysis, tumor 1, potentially characterized by myofibroblast-like malignant cells, could participate in facilitating tumor invasion and metastasis through its involvement in actin cytoskeleton reorganization (48). Tumor 2 could exhibit heightened proliferation and adhesion capabilities, suggesting its role as a subgroup primarily responsible for tumor growth (49). Tumor 3 could get involved in immune responses, while tumor 4 appeared to be a subgroup with a lower degree of differentiation. The latter three subgroups could collaborate in reshaping the non-cellular components of the TME, thereby fostering the progression of IFS (50). Additionally, when concentrating on CSCs (24), a distinct subset of self-renewable and highly proliferating neoplastic cells in solid tumor known as the ‘root cells’ of cancer, it was striking that three of four subtypes exhibited CSC phenotypes. This would partially elucidate the local high tumor invasiveness and CSCs might be new therapeutic targets for IFS. For example, cell-surface markers of CSCs such as CD133 and CD44 enable the targeted delivery of therapeutics such as chemotherapeutics and immuno-modulators (25).
When we explored the cell-cell communication landscape of TME in IFS, endothelial cells and macrophages were found to dominate the interaction network within this microenvironment. We identified multiple receptor-ligand interactions might contribute to tumor progression, highlighting their potential as therapeutic targets for IFS.
The vascular endothelial growth factor (VEGF) family could serve as an important role in tumorigenesis of IFS. VEGF family expressed by malignant cells stimulates the proliferation and survival of endothelial cells to initiate tumor neo-angiogenesis (26,27). VEGA+ malignant cells promote M2-like macrophage polarization to exert anti-inflammatory function (28). Also, M2-like macrophages express VEGA to promote tumor progression by the VEGFA/NRP-1/GAPVD1 axis (28). Currently, VEGF as a therapeutic target has been validated in various pediatric sarcomas (26,27). Considering the observed VEGF-related interactions above, it is worthwhile to explore the efficacy and safety of VEGF inhibitors in patients with IFS, especially those negative for the ETV6-NTRK3 fusion gene or resistant to NTRK inhibitors.
Besides, Jagged Canonical Notch Ligand (JAG), including JAG1 and JAG2, play a carcinogenic role in many cancer types (51,52). In IFS, JAG+ endothelial cells might enhance cancer stemness, aggressiveness and EMT of NOTCH+ malignant cells. We also found endothelial cells interact with malignant cells by JAG1/JAG2-VASN and VASN associated with tumor progression (53). The identification of JAG, previously unreported in IFS, presents a novel avenue for therapeutic intervention.
As widely identified in sarcomas (31), macrophages mainly display a M2-like phenotype in the TME of IFS. M2-like macrophages have been shown to predominantly interact with malignant cells, consequently promoting extracellular matrix remodeling and deposition, tumor angiogenesis, leukocyte recruitment, and immune suppression, all of which contribute to tumor progression (54). Numerous pro-tumorigenic interactions reported in other sarcomas, such as IL-34/CSF1-CSF1R and CD47-SIRPA (31,55), have also been observed between malignant cells and macrophages in IFS, thereby serving as potential immunotherapeutic targets. Furthermore, we identified some novel interactions, with limited or no previous reports in sarcomas but documented in other cancers, deserving further exploration. These include HBEGF-ERBB2/EGFR and CXCL2/CXCL10/CXCL12-DPP4 interactions (32-34). Our findings spotlight the potential of targeting macrophages for immunotherapy in IFS.
Our study had certain limitations that should be acknowledged. First, due to the rarity of IFS, the sample size was limited, consisting of only four samples, all of which were identified as having the ETV6-NTRK3 fusion gene. This lack of samples without the fusion gene may restrict the generalizability of our findings. Second, single-cell sequencing analysis is conducted based on bioinformatics methods and is limited to the mRNA level, requiring further experimental support and integration with multi-omics analysis to characterize tumor heterogeneity comprehensively. Our findings have not yet been validated through cellular and animal experiments. However, we are committed to conducting further research to supplement and confirm our initial findings in the future.
Conclusions
By conducting scRNA-seq, our study provided novel insights into the heterogeneity of IFS and characterized malignant cells and the TME at an unprecedented resolution. This high-resolution data presents an opportunity for making clinically significant advancements in the immunohistochemical diagnosis and treatment of IFS.
Acknowledgments
Funding: This work was funded by
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-24-66/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-24-66/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-24-66/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-24-66/coif). All authors report that this work was funded by Genertec Guozhong Healthcare (No. GZKJ-KJXX-QTHT-20220016), which is a for-profit company but has no conflicts of interest associated with this study. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Medical Ethics Committee of the Children’s Hospital of Fudan University [approval No. (2020). 419] and conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed written consent was obtained from the parents of the 23 included patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li Y, Dong R. Recent advances in the treatment of infantile fibrosarcoma. Chinese Journal of Pediatric Surgery 2020;41:1051-5.
- Davis JL, Al-Ibraheemi A, Rudzinski ER, et al. Mesenchymal neoplasms with NTRK and other kinase gene alterations. Histopathology 2022;80:4-18. [Crossref] [PubMed]
- Qi GW, Zheng J, Ma YY, et al. Clinicopathological analysis of infantile/congenital fibrosarcomas with rare histological features. Chinese Journal of Pathology 2019;48:700-4. [PubMed]
- Knezevich SR, McFadden DE, Tao W, et al. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet 1998;18:184-7. [Crossref] [PubMed]
- Albert CM, Davis JL, Federman N, et al. TRK Fusion Cancers in Children: A Clinical Review and Recommendations for Screening. J Clin Oncol 2019;37:513-24. [Crossref] [PubMed]
- Church AJ, Calicchio ML, Nardi V, et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod Pathol 2018;31:463-73. [Crossref] [PubMed]
- Wong V, Pavlick D, Brennan T, et al. Evaluation of a Congenital Infantile Fibrosarcoma by Comprehensive Genomic Profiling Reveals an LMNA-NTRK1 Gene Fusion Responsive to Crizotinib. J Natl Cancer Inst 2016;108:djv307. [Crossref] [PubMed]
- Laetsch TW, DuBois SG, Mascarenhas L, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol 2018;19:705-14. [Crossref] [PubMed]
- DuBois SG, Laetsch TW, Federman N, et al. The use of neoadjuvant larotrectinib in the management of children with locally advanced TRK fusion sarcomas. Cancer 2018;124:4241-7. [Crossref] [PubMed]
- Nagasubramanian R, Wei J, Gordon P, et al. Infantile Fibrosarcoma With NTRK3-ETV6 Fusion Successfully Treated With the Tropomyosin-Related Kinase Inhibitor LOXO-101. Pediatr Blood Cancer 2016;63:1468-70. [Crossref] [PubMed]
- Zhu H, Gu S, Yin M, et al. Analysis of infantile fibrosarcoma reveals extensive T-cell responses within tumors: Implications for immunotherapy. Pediatr Blood Cancer 2018; [Crossref] [PubMed]
- Lei Y, Tang R, Xu J, et al. Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol Oncol 2021;14:91. [Crossref] [PubMed]
- Lu T, Xu R, Wang C, et al. Bioinformatics analysis and single-cell RNA sequencing: elucidating the ubiquitination pathways and key enzymes in lung adenocarcinoma. J Thorac Dis 2023;15:3885-907. [Crossref] [PubMed]
- Awuah WA, Roy S, Tan JK, et al. Exploring the current landscape of single-cell RNA sequencing applications in gastric cancer research. J Cell Mol Med 2024;28:e18159. [Crossref] [PubMed]
- He S, Wang LH, Liu Y, et al. Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs. Genome Biol 2020;21:294. [Crossref] [PubMed]
- Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573-3587.e29. [Crossref] [PubMed]
- Wolock SL, Lopez R, Klein AM. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst 2019;8:281-291.e9. [Crossref] [PubMed]
- Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289-96. [Crossref] [PubMed]
- Li Z, Wang H, Dong R, et al. Single-Cell RNA-seq Reveals Characteristics of Malignant Cells and Immune Microenvironment in Subcutaneous Panniculitis-Like T-Cell Lymphoma. Front Oncol 2021;11:611580. [Crossref] [PubMed]
- Sun Y, Wu L, Zhong Y, et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 2021;184:404-421.e16. [Crossref] [PubMed]
- Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. [Crossref] [PubMed]
- Azizi E, Carr AJ, Plitas G, et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018;174:1293-1308.e36. [Crossref] [PubMed]
- Efremova M, Vento-Tormo M, Teichmann SA, et al. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484-506. [Crossref] [PubMed]
- Liu Y, Wang H. Biomarkers and targeted therapy for cancer stem cells. Trends Pharmacol Sci 2024;45:56-66. [Crossref] [PubMed]
- Chico MA, Mesas C, Doello K, et al. Cancer Stem Cells in Sarcomas: In Vitro Isolation and Role as Prognostic Markers: A Systematic Review. Cancers (Basel) 2023;15:2449. [Crossref] [PubMed]
- Nagl L, Horvath L, Pircher A, et al. Tumor Endothelial Cells (TECs) as Potential Immune Directors of the Tumor Microenvironment - New Findings and Future Perspectives. Front Cell Dev Biol 2020;8:766. [Crossref] [PubMed]
- Apte RS, Chen DS, Ferrara N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019;176:1248-64. [Crossref] [PubMed]
- Wang L, Zhang L, Zhao L, et al. VEGFA/NRP-1/GAPVD1 axis promotes progression and cancer stemness of triple-negative breast cancer by enhancing tumor cell-macrophage crosstalk. Int J Biol Sci 2024;20:446-63. [Crossref] [PubMed]
- Messex JK, Byrd CJ, Thomas MU, et al. Macrophages Cytokine Spp1 Increases Growth of Prostate Intraepithelial Neoplasia to Promote Prostate Tumor Progression. Int J Mol Sci 2022;23:4247. [Crossref] [PubMed]
- Landuzzi L, Ruzzi F, Pellegrini E, et al. IL-1 Family Members in Bone Sarcomas. Cells 2024;13:233. [Crossref] [PubMed]
- Zając AE, Czarnecka AM, Rutkowski P. The Role of Macrophages in Sarcoma Tumor Microenvironment and Treatment. Cancers (Basel) 2023;15:5294. [Crossref] [PubMed]
- Fu E, Liu T, Yu S, et al. M2 macrophages reduce the radiosensitivity of head and neck cancer by releasing HB-EGF. Oncol Rep 2020;44:698-710. [Crossref] [PubMed]
- Hu X, Wang X, Xue X. Therapeutic Perspectives of CD26 Inhibitors in Imune-Mediated Diseases. Molecules 2022;27:4498. [Crossref] [PubMed]
- Song N, Cui K, Zeng L, et al. Advance in the role of chemokines/chemokine receptors in carcinogenesis: Focus on pancreatic cancer. Eur J Pharmacol 2024;967:176357. [Crossref] [PubMed]
- Hu ZQ, Zhou SL, Zhou ZJ, et al. Overexpression of semaphorin 3A promotes tumor progression and predicts poor prognosis in hepatocellular carcinoma after curative resection. Oncotarget 2016;7:51733-46. [Crossref] [PubMed]
- Zhang D, Lindstrom A, Kim EJ, et al. SEMA3C Supports Pancreatic Cancer Progression by Regulating the Autophagy Process and Tumor Immune Microenvironment. Front Oncol 2022;12:890154. [Crossref] [PubMed]
- Ni B, Zhang D, Zhou H, et al. IL-34 attenuates acute T cell-mediated rejection following renal transplantation by upregulating M2 macrophages polarization. Heliyon 2024;10:e24028. [Crossref] [PubMed]
- Shao F, Gao Y, Wang W, et al. Silencing EGFR-upregulated expression of CD55 and CD59 activates the complement system and sensitizes lung cancer to checkpoint blockade. Nat Cancer 2022;3:1192-210. [Crossref] [PubMed]
- Wang D, Wang X, Si M, et al. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett 2020;474:36-52. [Crossref] [PubMed]
- Kihara S, Nehlsen-Cannarella N, Kirsch WM, et al. A comparative study of apoptosis and cell proliferation in infantile and adult fibrosarcomas. Am J Clin Pathol 1996;106:493-7. [Crossref] [PubMed]
- Bejarano L, Jordāo MJC, Joyce JA. Therapeutic Targeting of the Tumor Microenvironment. Cancer Discov 2021;11:933-59. [Crossref] [PubMed]
- Dorafshan S, Razmi M, Safaei S, et al. Periostin: biology and function in cancer. Cancer Cell Int 2022;22:315. [Crossref] [PubMed]
- Liu Y, Huang Z, Cui D, et al. The Multiaspect Functions of Periostin in Tumor Progression. Adv Exp Med Biol 2019;1132:125-36. [Crossref] [PubMed]
- Wei LF, Weng XF, Huang XC, et al. IGFBP2 in cancer: Pathological role and clinical significance Oncol Rep 2021;45:427-38. (Review). [Crossref] [PubMed]
- Zhang B, Hong CQ, Luo YH, et al. Prognostic value of IGFBP2 in various cancers: a systematic review and meta-analysis. Cancer Med 2022;11:3035-47. [Crossref] [PubMed]
- Liu YJ, Du J, Li J, et al. CTHRC1, a novel gene with multiple functions in physiology, disease and solid tumors Oncol Lett 2023;25:266. (Review). [Crossref] [PubMed]
- Mei D, Zhu Y, Zhang L, et al. The Role of CTHRC1 in Regulation of Multiple Signaling and Tumor Progression and Metastasis. Mediators Inflamm 2020;2020:9578701. [Crossref] [PubMed]
- Izdebska M, Zielińska W, Hałas-Wiśniewska M, et al. Involvement of Actin and Actin-Binding Proteins in Carcinogenesis. Cells 2020;9:2245. [Crossref] [PubMed]
- Yayan J, Franke KJ, Berger M, et al. Adhesion, metastasis, and inhibition of cancer cells: a comprehensive review. Mol Biol Rep 2024;51:165. [Crossref] [PubMed]
- Sala M, Ros M, Saltel F. A Complex and Evolutive Character: Two Face Aspects of ECM in Tumor Progression. Front Oncol 2020;10:1620. [Crossref] [PubMed]
- Meurette O, Mehlen P. Notch Signaling in the Tumor Microenvironment. Cancer Cell 2018;34:536-48. [Crossref] [PubMed]
- Qiao X, Ma B, Sun W, et al. JAG1 is associated with the prognosis and metastasis in breast cancer. Sci Rep 2022;12:21986. [Crossref] [PubMed]
- Wan F, Li H, Huang S, et al. Vasorin promotes proliferation and migration via STAT3 signaling and acts as a promising therapeutic target of hepatocellular carcinoma. Cell Signal 2023;110:110809. [Crossref] [PubMed]
- Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010;141:39-51. [Crossref] [PubMed]
- Chang WI, Lin C, Liguori N, et al. Molecular Targets for Novel Therapeutics in Pediatric Fusion-Positive Non-CNS Solid Tumors. Front Pharmacol 2021;12:747895. [Crossref] [PubMed]


